PASS
Our Pulmonary Analysis Software Suite has been developed by in house programmers. It is a comprehensive package for the manipulation, display and analysis of multidimensional image data sets. PASS is intended to assist doctors and researchers in the field of pulmonary research with lung segmentation, histogram analysis, hole measurement, and nodules and tissue classification.
Apollo
As VIDA's organically-developed software application, Apollo®, has evolved for over 10 years with extensive validation in marquis clinical trials and research institutions. Apollo has significant algorithmic, workflow and automation features applicable to high volume, repeatable parenchymal, airway, and fissure integrity analysis. VIDA believes that understanding the analysis software to its very core is imperative for high quality so they only use their own applications
MIFAR
Medical Imaging File Archive Recovery - MIFAR - is a set of software tools for storing medical image data along with other associated post-image processing files, physiologic records, and lab notes.
PVS
Procedural Verification Software is an automated web portal system for tracking and verifying data for multi-center studies. It provides a mechanism for obtaining information from scans including deviations and scan parameters as well as the appropriate dose for a given visit. PVS users are able to register subjects for their site in the database, communicate with the Radiology Reading Center via messaging system, and view details of previous scans performed by their site.
TSIA
Perform perfusion and ventilation (blood-flow) analysis. Developed within our lab. The name stands for Time Series Image Analysis. It is cross-platform so it functions on Windows, Mac OSX, and Linux. The concept of TSIA was developed in the early 1990's. Algorithm modules for perfusion/ventilation parameter calculation are separated from GUI and they are built as a dynamic link library. Therefore, any algorithm dll can be switched depending on the data set at runtime. This enables algorithm developers to easily create their own algorithms for your analysis.
Facilities
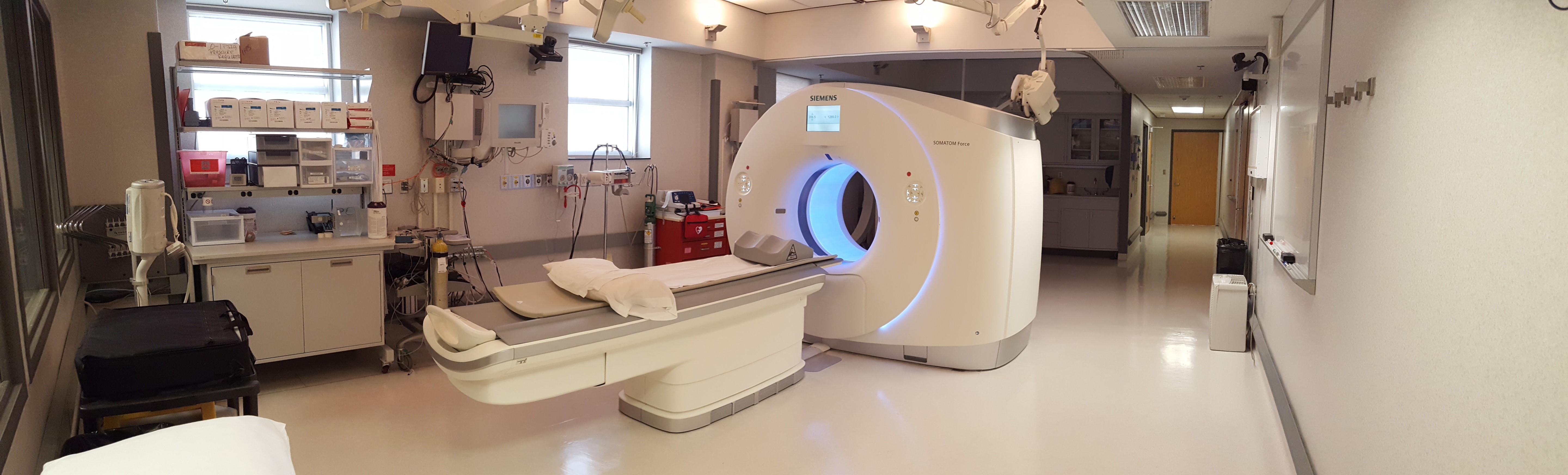
The University of Iowa Carver College of Medicine houses a 2500 square foot CT imaging research facility for the support of the research efforts of the Advanced Pulmonary Physiomic Imaging Laboratory, APPIL. This space is located strategically between the patient areas of the University of Iowa Hospitals and Clinics, the NIH supported Clinical Research Unit, and the Animal Care Facilities of the College of Medicine. The space has been designed with a large control room, a MDCT scanner suite, a human preparation area, an animal preparation area, and a micro-CT room with a separate control room adjacent. The CT Scanning Research Facility is equipped with Polycom cameras within the CT suite, CT control room, animal surgery suite, and micro CT suite. It has a Polycom Server that allows multi-point conferencing and the ability for any of the remote sites to participate in live experiments. A Philips physiological monitoring system has been installed to match that in the Medical Intensive Care Unit in the UI Hospital. Data from the physiologic system is fed to a monitoring system running LabView software that interprets the physiology and sends signals to the MDCT scanner such that images are captured at a precise point within the physiologic signal of interest. The CT control room has a 15ft observation window into the CT suite, a conference table and 7 computer equipped carrels for visiting scientists, the CT technologist and a study coordinator. Other major equipment include a Xe gas re-breathing delivery system (Diversified Diagnostic Products, Windfern, TX), high pressure, dual head MedRad contrast injector, a dual headed Bracco contrast injector, and a mobile digital fluoroscopy unit, allowing catheterization procedures to be performed in the human and animal preparation suites or at the scanner table. The facility includes a custom-built dual piston system that controls inspiration and expiration during human scanning such that exactly similar volumes of gas (xenon mixture) and exhalate are inspired from and exhaled into the two pistons with valves under computer control. It also includes a custom-built volume controller system using LabView software, which allows us to monitor human breathing in order to reach precise lung volumes for the scan.
- Dual Source Multiple Detector Computed Tomography Scanner: With our relationship with Siemens, we originally installed a 16 slice MDCT scanner which was upgraded to a Sensation 64 and later to the 128 slice Definition Flash. The Definition Flash was a dual source, dual energy CT scanner with Stellar detectors and Safire iterative reconstruction option. On July 2015, the lab was upgraded to a SIEMENS SOMATOM Force Dual Source CT. This new scanner allows for a reduced dose of up to 50% compared to other modern CT systems. This is in conjunction with a considerable increase in sensitivity and specificity associated with Dual Source DE scanning. It is a 384 (2 X 192) slice scanner with speeds of 0.25 seconds per rotation. Additionally, the Force has unprecedented power reserves in the form of two 120 kW generators that enable scanning of patients at low kV from 70 to 90 kV.
- MicroCT Scanner:
 The Imtek (now Siemens) CAT II micro CT scanner (purchased under an NIH SIG) has the capability of imaging a mouse in vivo with approximately 20µ resolution in approximately 20 minutes. Jessica Sieren, PhD, has recently been awarded funding to add a ZEISS Xradia 520 Versa submicron 3D X-ray CT scanner that has been installed.
The Imtek (now Siemens) CAT II micro CT scanner (purchased under an NIH SIG) has the capability of imaging a mouse in vivo with approximately 20µ resolution in approximately 20 minutes. Jessica Sieren, PhD, has recently been awarded funding to add a ZEISS Xradia 520 Versa submicron 3D X-ray CT scanner that has been installed. - 3T Premier MRI Scanner:
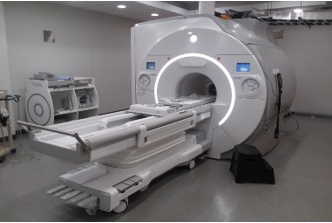 As of Feb 2020, APPIL has access to the new GE 750W Premier system housed in the Iowa Institution for Biomedical Imaging. This is a state of the art 3T MRI scanner which has been customized by the vendor to allow imaging on the Xenon channel. High gradient amplitude of 8 G/cm in each plane allows for rapid and effective diffusion weighted imaging. This system is fully compatible with hyperpolarized noble gas MRI performed for pulmonary ventilation imaging, as well as imaging on the MRI proton channel for structural imaging and oxygen-enhanced lung imaging.
As of Feb 2020, APPIL has access to the new GE 750W Premier system housed in the Iowa Institution for Biomedical Imaging. This is a state of the art 3T MRI scanner which has been customized by the vendor to allow imaging on the Xenon channel. High gradient amplitude of 8 G/cm in each plane allows for rapid and effective diffusion weighted imaging. This system is fully compatible with hyperpolarized noble gas MRI performed for pulmonary ventilation imaging, as well as imaging on the MRI proton channel for structural imaging and oxygen-enhanced lung imaging. - Polarean 9820 Xenon Polarization System:
 APPIL was the recent beneficiary of a shared instrumentation award to obtain a Polarean 9820 Senon polarizer. This is a dedicated system for the purpose of polarization of Xenon gas to be used for the assessment of regional lung structure and function via MRI. Throughput is approximately 3L of gas per hour at 40% polarization. In conjunction with the Premier 3T MRI scanner, this system allows lung ventilation imaging, static and dynamic, with Xenon, as well as diffusion weighted imaging and gas transfer mapping.
APPIL was the recent beneficiary of a shared instrumentation award to obtain a Polarean 9820 Senon polarizer. This is a dedicated system for the purpose of polarization of Xenon gas to be used for the assessment of regional lung structure and function via MRI. Throughput is approximately 3L of gas per hour at 40% polarization. In conjunction with the Premier 3T MRI scanner, this system allows lung ventilation imaging, static and dynamic, with Xenon, as well as diffusion weighted imaging and gas transfer mapping.
In addition to this space, there is separate new space immediately above for housing our computer clusters, large array data storage racks. Next to this room is a dedicated fully equipped pulmonary function laboratory including a body box (Nspire) and a gas chromatograph, allowing correlative studies using the Multiple Inert Gas Elimination Technique (MIGET). Directly opposite the computer facility is the Environmental Health Sciences Research Center Exposure Chamber Facility with associated pulmonary function lab. Dr Hoffman is the director of this facility. On this floor level and 2 floors above this floor resides the NIH supported CTSA Clinical Research Center. In addition, the I-Clic has a customized microscopy facility with what has been dubbed the LIMA system consisting of a large stage microtome and microscope linked by computer control such that the cut surface of a specimen is digitized at microscopic resolution, the surface is scanned and images are merged before a thin section is cut, the microtome stage is adjusted such that the new surface remains in focus and the process is repeated. Dr. Hoffman’s 2,500 square foot office and image analysis space reside on the 7th floor above the scanner facility.
CFD (Computational Fluid Dynamic Software)
Multiscale imaging-based lung model suite (MSILung) is an integrative computational framework that comprises a multi-scale geometric modeling code. MSILung is multi-scale in three aspects:
- bridging central and peripheral airways and linking structure and function
- bridging organ and cellular (or molecular) scales
- bridging individual and population scales
MSILung allows assessment and prediction of lung function for improved patient phenotyping and hence patient-specific therapy. Our in-house CFD code is based on a characteristic-Galerkin finite element method (FEM) for simulation of three-dimensional (3D) gas flow in complex geometries. The CFD thermodynamics model can predict evaporation and condensation of water vapor on the airway wall without prescribing wall temperature a priori. Our in-house multi- scale geometric human airway model allows automatic construction of subject-specific 3D airway geometrical model and CFD mesh. To model airways with moving walls as in a breathing lung, we adopt the Arbitrary-Lagrangian-Eulerian (ALE) approach. To capture accurately the flow in various turbulent, transient and laminar regimes in the lungs, large-eddy simulation (LES) technique is adopted. Our image-based technique provides a physiologically consistent subject-specific flow boundary condition (BC) for CFD. The regional ventilation information is derived by our in-house mass-preserving image registration of CT lung images acquired at two inflation levels. Image registration not only provides BCs and deforming airway geometries for CFD analysis, but also derives regional functional variables for cluster analysis. The above CFD and BC methods can be applied to multiple images, e.g. three static images and 4D-CT images, for a breathing lung simulation. For particle simulations in the study of drug delivery and environmental factors (e.g. inhaled pollutants), we adopt the Lagrangian tracking algorithm to compute particle trajectories and depositions. Our CFD code is parallelized with Message-Passage Interface (MPI), and is executed on 500-1000 CPU cores with almost linear scalability. Our GPU-based image registration code is based on as computation- and memory-efficient Diffeomorphic Mulit-level B-spline Transform Composite (DMTC) method on GPU for performing nonrigid mass-preserving registration of two CT volumetric lung images. The method is based upon the sum of squared local tissue volume differences (SSTVD) similarity criterion to preserve lung tissue mass. The registration code components modified for GPU computation demonstrated speedup of 112 times faster than a serial CPU version and 11 times that of a 12- threaded CPU version.
Interested in using our facility?
For questions and scheduling contact Jarron Atha:
- jarron-atha@uiowa.edu
- Phone: 319-384-7946
- Fax: 319-384-7929